May 29, 2015
We have a new publication in ISME Journal that discusses the potential role of diatom proteorhodopsins in helping cope with low iron availability. Co-authors include Dylan Catlett (former UNC undergraduate research scientist), Kelsey Ellis, Brian Hopkinson from UGA and Nicolas Cassar from Duke University.
To read the full article, go here.
Our study was also highlighted on the UNC College of Arts and Sciences webpage and can be found here.
December 8, 2014
Our very own Kelsey Ellis published a piece in the October issue of Natural History magazine about conducting research on diatoms. You can read it here.
p.s. Prospective students, don’t be dissuaded by what Kelsey describes as the “monotony” of phytoplankton research. In fact, what research areas aren’t monotonous at times? And how many of these provide you with the opportunity to join research cruises to far-off, interesting places?
August 1, 2014
We recently received NSF funding to investigate the effects of iron and light limitation within ecologically important polar diatoms isolated from the Southern Ocean. This is part of a collaboration with Dr. Nicolas Cassar’s group at Duke University.
More information about this award can be found here.
July 6, 2014
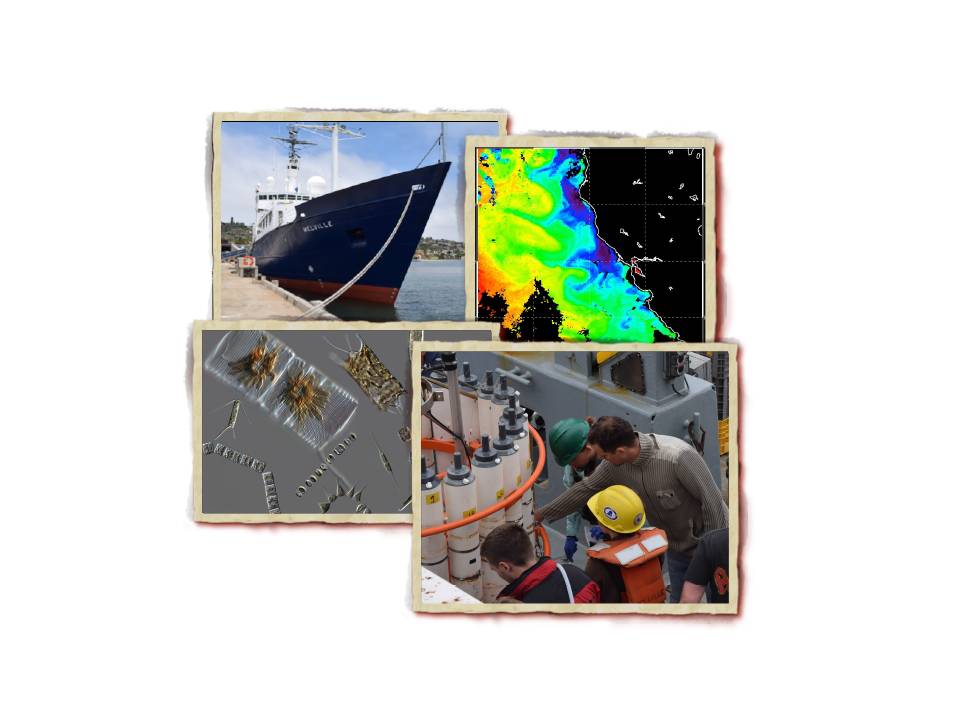 The Marchetti Lab is participating on a research cruise along the Caifornia coast to investigate the ecological importance of iron storage in diatoms aboard the R/V Melville during July 2014 as part of an NSF funded project. Miriam Sutton, a middle school science teacher has joined the Marchetti group to convey her experiences through her classroom website: Science by the Sea. You can follow Miriam’s adventures at sea through her website blog or through her “Science by the Sea” social networks, including Facebook, Twitter, and Instagram.
The Marchetti Lab is participating on a research cruise along the Caifornia coast to investigate the ecological importance of iron storage in diatoms aboard the R/V Melville during July 2014 as part of an NSF funded project. Miriam Sutton, a middle school science teacher has joined the Marchetti group to convey her experiences through her classroom website: Science by the Sea. You can follow Miriam’s adventures at sea through her website blog or through her “Science by the Sea” social networks, including Facebook, Twitter, and Instagram.
August 1, 2013
We recently received NSF funding to investigate the ecological role of iron storage in diatoms. This is a collaborative research project with Dr. Benjamin Twining’s group at the Bigelow Laboratory for Ocean Sciences.
More information about this award can be found here.
December 13, 2012
The research conducted in our lab was featured in the Fall issue of Carolina Scientific, a UNC scientific magazine written, edited and designed by UNC undergraduate students that features the cool science conducted at UNC – Chapel Hill. An article written by Hannah Alchelman, who is completing a minor in Marine Sciences, that highlights our research is on Page 22. We also made the front cover!

October 6, 2012
Our study investigating how variable iron states influences the precipitation of silica in diatoms was published this week in L&O. The main findings of this study are that diatom composition influences the overall community silica precipitation much more than their iron status and that diatoms use different silicic acid transporters when they are iron-limited versus when they are iron replete. Colleen Durkin is the lead author of this study.
To read the full article, go here.
To learn more about Colleen’s research, go here.
October 2, 2012
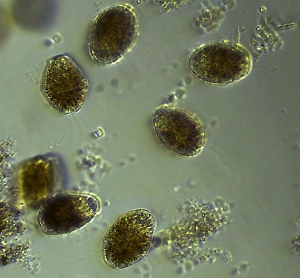
The dinoflagellate cf. Gyrodinium within a water sample collected from the Neuse RIver Estuary
Last week there were numerous harmful algal blooms in the Neuse River Estuary (NRE) associated with some extensive fish kills. Fortunate for us (but not so fortunate for the Menhaden), we sampled a bloom of the dinoflagellate cf. Gyroodinium for metatranscriptomic sequencing as part of a project to evaluate the usefulness in integrating high-throughput sequencing approaches to assess the taxonomy and molecular physiology of eukaryotic phytoplankton in the NRE. This project is funded by the NC Sea Grant and is a collaboration between the Marchetti lab and Hans Paerl’s lab at the Institute of Marine Sciences in Morehead City.
January 19, 2012
Our study titled ” Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability” was published in the Proceedings of the National Academy of Science. To our knowledge this is the first study to implement high-throughput sequencing techniques to assess how marine eukaryotic plankton respond to environmental change.
A link to the PNAS webpage is here.
A PDF of the publication is here.
A PDF of the Author Summary is here
November 15, 2011
Next-generation (and now third generation) sequencing technologies are enabling molecular biologists to obtain massive sequence libraries of DNA and RNA directly from the environment. The ability to determine the taxonomic affiliation of a sequence read along with the gene it likely encodes necessitates an extensive reference sequence database of organisms. From marine environments, there are upwards of hundreds of prokaryotic genomes sequenced providing a fairly extensive reference genome database (although I must acknowledge that marine bacteria and archaea are incredibly diverse). For marine eukaryotes, including both autotrophic and heterotrophic members, our reference sequence databases are very sparse, for the most part, due to their larger genomes and difficulty to maintain in culture. Just recently, genomes and extensive Expressed Sequence Tag (EST) libraries for members from each of the major marine eukaryotic phytoplankton function groups have been sequenced. This includes representatives of diatoms, coccolithophores, dinoflagellates, green algae and cryptomonads. However these currently available reference databases for marine eukaryotes represent only a very minute fraction of the total species that exist within marine environments. This creates a road block for taxonomic identification and functional gene annotation of reads in metagenomic and metatranscriptomic projects from marine environments (see my research page for an example of such a study). With the aims of providing an increased ability for the interpretation of marine sequencing projects the Gordon and Betty Moore Foundation are funding sequencing of 750 EST libraries of marine microeukaryotes using the Illumina HTSeq 2000 Platform. Sequencing is currently underway. I am pretty excited about this large sequencing effort, although I wish they would have initiated this sooner!
More details about this project can be found here.




University Operator: (919) 962-2211 | © 2024 The University of North Carolina at Chapel Hill |