Marchetti Lab
ecophysiology, biogeochemistry and genomics of marine phytoplankton
Marchetti Lab Research Themes:
- Trace metal and vitamin availability controls on phytoplankton ecology and evolution
- Diatom elemental requirements and nutrient uptake kinetics
- Comparative transcriptomics in phytoplankton isolates and natural assemblages
- Development of molecular and morphological indicators for phytoplankton physiological status
- Phytoplankton-bacteria interactions
- Climate change effects on phytoplankton and zooxanthellae
To view a map of field sites where we are currently conducting our research, click here.
Below are some ongoing and completed research projects:
 The influence of different nutrient delivery modes on functional biodiversity of marine plankton in a changing ocean
The influence of different nutrient delivery modes on functional biodiversity of marine plankton in a changing ocean
All living organisms must acquire nutrients from their environment to survive and grow. Phytoplankton and their zooplankton grazers in the ocean, which constitute the base levels of the planet’s largest food webs and play an essential role in global carbon cycling, are no exception. However, the rules governing how different physical mechanisms of nutrient delivery to marine ecosystems structure plankton biodiversity and interaction networks along with their trophic dependencies are poorly understood. The overarching goal of the project is to examine how these specific physical mechanisms influence the functional biodiversity of plankton communities through increasing nutrient inputs into the mixed layer, leading to strong, yet perhaps predictable biological variability in a changing ocean environment. The project will combine field and laboratory studies with a modeling effort to evaluate if different physical modes of nutrient delivery, in close proximity to each other, exert sufficient controls on nutrient availability that influences plankton functional diversity and associated dynamics. The study site is the Galápagos archipelago in the eastern equatorial Pacific Ocean. Intensive shipboard sampling will be augmented by 12-month deployments of a vertical profiler mooring to observe i) an isolated island wake and ii) interaction of the archipelago with the Equatorial Undercurrent (EUC) and upwelling on the western side of the archipelago. Internal tides are believed to have a significant influence on the system and also will be observed. This research will advance ecological theory on biodiversity dynamics and functional biodiversity by testing the hypothesis that functional redundancy drives ecosystem stability, achieved through resistance to change or resiliency.
Funded by NSF Biodiversity on a Changing Planet program (in collaboration with H. Seim from UNC, M. Messié from MBARI and D. Figueroa from UT-RGV)
 Export Processes in the Ocean from Remote Sensing (NASA EXPORTS)
Export Processes in the Ocean from Remote Sensing (NASA EXPORTS)
As part of the EXPORTS program, we sought to quantify and relate primary productivity, remineralization and net community production (NCP) in the mixed layer across different ecosystem/carbon cycling states (ECCs). More specifically, our measurements contributed towards the determination of upper ocean ecosystem characteristics that are important in controlling the vertical transfer of organic matter to ocean depths. Through a combination of biogenic O2 gas and inorganic carbon and nitrogen inventory measurements, we quantified the biological rate processes and fluxes in both autotrophic and heterotrophic members of the marine microbial community. Estimates of Gross Primary Productivity (GPP) and respiration rates in conjunction with mixed-layer integrated rates of NCP were measured to quantify the overall carbon export from the mixed layer. In addition to the composition of the microbial community present (i.e., phytoplankton, bacteria and archaea) there is increasing evidence that their physiological status is also important in predicting carbon export. Thus, in tandem with our rate measurements, we determined the marine microbial community composition through targeted DNA sequencing as well as sequencing of environmental RNA to determine gene expression that can be used to infer the physiological status of the microbial plankton community. Together, the integration of these measurements will allow for an unprecedented ability to examine how the form and function of upper ocean microbial ecosystems shape the carbon export potential across different ECCs.
Funded by NASA (in collaboration with S. Gifford from UNC and N. Cassar from Duke)
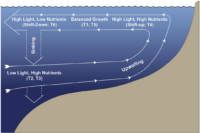 Phytoplankton response to the UPwelling Conveyor Belt CYCLE (PUPCYCLE)
Phytoplankton response to the UPwelling Conveyor Belt CYCLE (PUPCYCLE)
Upwelling zones are hotspots of primary productivity that are characterized by having strong spatial and temporal dynamics. Phytoplankton bloom when deep, nutrient-rich waters are upwelled into sunlit surface layers of the ocean, providing nourishment that supports productive food webs and extensive drawdown of carbon dioxide from the atmosphere to ocean depths. These photosynthetic microbes must constantly acclimate to changes in their chemical and physical environments, as subsurface seed populations shift up their metabolisms in response to changes in light conditions associated with upwelling. When upwelled waters age, cells sink out of the photic zone, establishing seed populations that remain dormant until the next upwelling event; a process referred to as the upwelling conveyor belt cycle (UCBC). Our research examines the following questions: a) How do phytoplankton respond at the molecular and physiological level to the different UCBC stages, b) which seed populations (i.e., surface versus subsurface) contribute most to phytoplankton blooms during a strong upwelling event, c) how are phytoplankton elemental compositions altered throughout UCBC stages, and d) how do iron limitation affect the phytoplankton responses to UCBC conditions? This research will combine field and lab work where cruises in the California Upwelling Zone (CUZ) will study the dynamics of phytoplankton communities within natural upwelled water and simulated upwelling incubation experiments. In the lab, the transcriptional responses and changes in elemental composition of phytoplankton isolates exposed to UCBC-like conditions will be examined in order to test mechanistic hypotheses and assess biogeochemical feedbacks.
Funded by NSF Biological Oceanography Programthrough a CAREER award to A. Marchetti
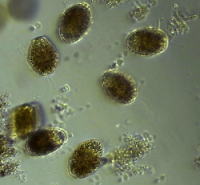
The dinoflagellate Levanderina sp. within a seawater sample collected in the Neuse River Estuary, NC.
Integrating Eukaryotic Plankton Community Transcriptomics into an Ecological Network Analysis of the Neuse River Estuary
Eutrophication of estuarine and coastal systems can often result in harmful algal blooms (HABs), hypoxia and fish kills. Non-point sources of pollution in the Neuse River Estuary (NRE) watershed have been increasing as the area has experienced steady growth in agriculture, industry and urbanization. Efforts to reduce nutrient inputs of phosphorus have had positive results; however, similar efforts for nitrogen input reduction have not been as successful. Necessary to the effective management of this ecosystem are the identification of the abiotic and biotic components of the system and subsequent understanding of the relationships of those components. The aim of this project is to provide insight into bloom dynamics, causes, and effects, and to provide new molecular tools that may aid in forecasting HABs. We seek to build ecological relationships between the plankton communities incorporating transcriptomic (analysis of RNA sequences) data from eukaryotic phytoplankton. The utilization of next generation sequencing technologies will provide high-resolution of biotic components of this ecosystem. These data will be contextualized with environmental data routinely collected by the ModMon monitoring program in the NRE, such as chlorophyll a, particulate organic matter, primary productivity and dissolved organic and inorganic nutrients. The coupling of these data, with an emphasis on nitrogen cycling, will elucidate important ecological networks that can then be used to guide best management practices to promote healthy ecosystems. The models, approaches, and tools produced by this study could be expandable to other coastal and estuarine systems.
Funded by North Carolina Sea Grant (Collaborative proposal with Dr. Hans Paerl from UNC Institute of Ocean Sciences)
Gong W, Browne J, Hall N, Schruth D, Paerl H, and Marchetti A. (2016) Molecular insights into a dinoflagellate bloom. ISME Journal. doi:10.1038/ismej.2016.129
Additional links discussing this project:
UNC Endeavors – Sequencing the Sea
Data archive can be found here.
Data archive can be found here.
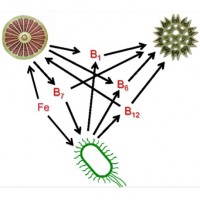
Iron and Vitamin Interactions in Diatoms
Iron and B-vitamins can play important roles in regulating the growth of bacteria and protist phytoplankton in our oceans, yet it is not well understood how they respond when faced with simultaneous iron and vitamin stress. Although most phytoplankton, including diatoms, can synthesize some B-vitamins (e.g. biotin [B7]), other vitamins may only be supplied to phytoplankton through bacterial production (e.g. cobalamin [B12]), possibly resulting in vitamin exchanges between these microorganisms. This study aims to understand the interactions between iron and B-vitamin limitation within laboratory diatom cultures and natural phytoplankton assemblages along a natural iron gradient in the Northeast Pacific Ocean. The pairing of complementary approaches including physiological measurements, dissolved B-vitamin concentrations, and community transcriptomics provides us with a better understanding of phytoplankton responses to variations in iron and vitamin availability, which ultimately may be controlling plankton community composition, primary production, and nutrient cycling in this region as well as in other iron-limited regions throughout the globe.
Data archive can be found here.
Data archive can be found here.
 Investigating an Ecological Role for Iron Storage in Diatoms
Investigating an Ecological Role for Iron Storage in Diatoms
A fundamental goal in phytoplankton ecology is to identify environmental factors that control phytoplankton distribution and abundance. In 30-40% of the world’s oceans, phytoplankton growth is limited by availability of the micronutrient iron. Addition of iron to surface waters in these regions induces massive phytoplankton blooms dominated primarily by large, mostly pennate diatoms. This suggests that the diatoms able to bloom in response to these sporadic iron deliveries are able to rapidly take up and store iron, permitting continued divisions once the chronically low iron concentrations return. Many animals, plants and bacteria store iron through highly specialized iron storage proteins known as ferritins. Recently, ferritin-encoding genes were discovered within the bloom-forming pennate diatoms, marking the first description of ferritin in any member of the Stramenopiles, a diverse Eukaryotic lineage that includes unicellular algae, macroalgae and plant parasites. However, it is not known whether there are systematic differences in iron-storage capacities between ferritin-containing diatoms and those that lack ferritin. The overall objective of this project is to examine the ecological importance of iron storage as a selective mechanism controlling the distributions of diatoms along natural and artificial iron gradients in oceanic and coastal marine ecosystems. Through a combination of laboratory and field studies we will determine the ability of oceanic and coastal diatoms that either contain or lack ferritin to perform luxury uptake of iron and utilize these iron stores in the absence of an external source of iron to maintain growth and out compete other members of the phytoplankton assemblage. In the laboratory we are investigating the iron storage capacities within a variety of ferritin containing and non-containing oceanic and coastal diatoms. In the field we are partaking on two cruises, one to the California Current System in July of 2014 to examine iron storage capacities in coastal diatom communities and the other to Ocean Station Papa in the Northeast Pacific Ocean in June of 2015 to evaluate iron storage capacities in oceanic diatom communities.
Funded by NSF Biological Oceanography Program (Collaborative proposal with Dr. Ben Twining from the Bigelow Laboratory of Ocean Sciences)
Iron and Light Limitation in Southern Ocean Diatoms: Comparative Transcriptomics and Molecular Indicators
The Southern Ocean surrounding Antarctica is changing rapidly in response to Earth’s warming climate. These changes will undoubtedly influence communities of primary producers by altering conditions that influence their growth and composition. Because primary producers such as phytoplankton play an important role in global biogeochemical cycling, it is essential to understand how they will respond to changes in their environment. The growth of phytoplankton in certain regions of the Southern Ocean is constrained by steep gradients in chemical and physical properties that vary in both space and time. Light and iron have been identified as key variables influencing phytoplankton abundance and distribution within Antarctic waters. Microscopic algae known as diatoms are dominant members of the phytoplankton and sea ice communities, accounting for significant proportions of primary production. The overall objective of this project is to identify the molecular bases for the physiological responses of polar diatoms to varying light and iron conditions. The project should provide a means of evaluating the extent these factors regulate diatom growth and influence net community productivity in Antarctic waters. Although numerous studies have investigated how polar diatoms are affected by varying light and iron, the cellular mechanisms leading to their distinct physiological responses remain unknown. Using comparative transcriptomics, the expression patterns of key genes and metabolic pathways in several ecologically important polar diatoms recently isolated from Antarctic waters and grown under varying iron and irradiance conditions will be examined. In addition, molecular indicators for iron and light limitation will be developed within these polar diatoms through the identification of iron- and light-responsive genes — the expression patterns of which can be used to determine their physiological status. Upon verification in laboratory cultures, these indicators will be utilized by way of metatranscriptomic sequencing to examine iron and light limitation in natural diatom assemblages collected along environmental gradients in Western Antarctic Peninsula waters. In order to fully understand the role phytoplankton play in Southern Ocean biogeochemical cycles, dependable methods that provide a means of elucidating the physiological status of phytoplankton at any given time and location are essential.
Funded by NSF Antarctic Organisms and Ecosystems Program (Collaborative project with Dr. Nicolas Cassar from Duke University)
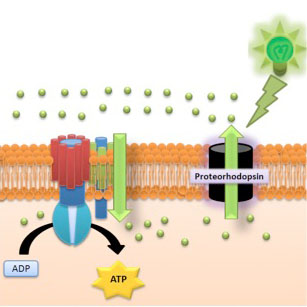 Examining the functional roles of proteorhodopsins in marine diatoms
Examining the functional roles of proteorhodopsins in marine diatoms
Proteorhodopsins (PR) are retinal-binding membrane proteins that function as light-driven proton pumps to generate energy for metabolism and growth. Recently PR-like genes have been identified in some marine eukaryotic protists, including diatoms, dinoflagellates, haptophytes and cryptophytes. These rhodopsins are homologous to green-light absorbing, ATP-generating PRs present within bacteria. In the oceanic diatom Pseudo-nitzschia granii, PR-like gene and protein expressions increase appreciably under iron limitation. In a survey of available transcriptomes, PR-like genes in diatoms are generally found in isolates from marine habitats where seasonal to chronic growth limitation by the micronutrient iron is prevalent, yet similar biogeographical patterns are not apparent in other phytoplankton taxa. We therefore propose that rhodopsin-based phototrophy could account for a proportion of energy synthesis in marine eukaryotic photoautotrophs, especially when photosynthesis is compromised by low iron availability. We are now investigating whether proteorhodopsins can account for a significant proportion of energy production in diatoms and whether this energy would allow them to fix carbon dioxide. This alternative ATP-generating pathway could have significant effects on plankton community structure and global ocean carbon cycling
Funded by NSF Antarctic Organisms and Ecosystems Program
 Transcriptomic responses of coral hosts and their symbionts to thermal and ocean acidification stressors
Transcriptomic responses of coral hosts and their symbionts to thermal and ocean acidification stressors
Elucidating the responses of symbiotic partners to environmental stressors is becoming increasingly important as climate change disrupts biotic interactions worldwide. We exposed the resilient and ubiquitous Caribbean coral Siderastrea siderea and its Clade C symbiont Symbiodinium to temperature and pCO2 treatments for 95 days and quantified the transcriptomic responses of each partner using RNAseq. Both elevated temperature and pCO2 elicited strong but divergent responses of the host’s transcriptome. High temperatures disrupted molecular homeostasis and substantially reduced calcification rates. Conversely, elevated pCO2 enhanced host transcription of genes associated with respiration and hydrogen ion transport, with only minimal effects on calcification rates—underscoring the role of proton transport in calcification maintenance under elevated pCO2, while also suggesting costs associated with this acclimation. Contrary to the host, the symbiont’s transcriptome exhibited little change in response to elevated pCO2. Instead, population-specific transcriptomic responses were observed across fore-reef and near-shore environments—consistent with observed differences in symbiont photosynthetic efficiency across these two reefs. We conclude that host transcriptomic plasticity promotes acclimation to ocean acidification, but not necessarily warming. Given the host’s strong transcriptomic responses to acidification and warming, coupled with the symbiont’s lack of response, we hypothesize that hosts actively buffer their symbiont’s environment.
Siderastrea siderea transcriptome: Assembled and annotated transcriptome for the adult scleractinian coral S. siderea with symbiont contamination removed. Data were generated using Illumina HiSeq2000 2*100bp reads and assembled using Trinity. Link to transcriptome
Davies SW, Ries JB, Marchetti A and Castillo DK. (2018) Symbiodinium Functional Diversity in the Coral Siderastrea siderea Is Influenced by Thermal Stress and Reef Environment, but Not Ocean Acidification. Frontiers in Marine Science: doi.org/10.3389/fmars.2018.00150
Clade C Symbiodinium transcriptome: Assembled and annotated transcriptome for the symbiotic dinoflagellate algae Clade C Symbiodinium hosted by Siderastrea siderea with all host contamination removed. Data were generated using Illumina HiSeq2000 2*100bp reads and assembled using Trinity. Link to transcriptome
 Comparative transcriptomics provides insights into the iron-related architecture of an oceanic diatom
Comparative transcriptomics provides insights into the iron-related architecture of an oceanic diatom
Oceanic diatoms are well adapted to growing at low iron concentrations that persist in offshore surface waters. Their rates of carbon dioxide fixation per unit of iron used, referred to as the iron-use efficiency, are much higher than their coastal counterparts, thus reducing their demand for iron relative to the supply. Previously, this streamlining of iron requirements had mostly been assessed at the level of single genes, proteins or pathways. We performed a whole-cell comparative transcriptomic analysis of the oceanic pennate diatom Pseudo-nitzschia granii isolated from iron-poor waters of the NE Pacific Ocean. Using a combination of next-generation sequencing (NGS) techniques the transcriptomes of Fe-limited cells and those recently resupplied with Fe were compared. Based on our statistical approach, of the 4800 predicted genes identified in the transcriptomes, 2709 genes were significantly differentially expressed (p<0.05) alluding to a substantial reorganization of the cellular constituents as a consequence of Fe-limited growth. The comparative transcriptome of P. granii establishes proof of concept for the successful implementation of NGS in evaluating whole-cell differential expression and provides a mechanistic basis for the exceptional ability of oceanic diatoms to survive in low iron environments.
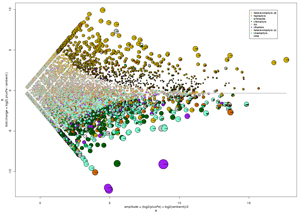
Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability
In vast expanses of the oceans, growth of large phytoplankton such as diatoms is limited by iron availability. Diatoms respond almost immediately to the delivery of iron and rapidly comprise the majority of phytoplankton biomass. The molecular bases underlying the subsistence of diatoms in iron-poor waters and the plankton community dynamics that follow iron-resupply remain largely unknown. Here we use comparative metatranscriptomics to identify changes in gene expression associated with iron-stimulated growth of diatoms and other eukaryotic plankton. A microcosm iron-enrichment experiment using mixed-layer waters from the NE Pacific Ocean resulted in increased proportions of diatom transcripts and reduced proportions of transcripts from most other taxa within 98 hours after iron addition. Hundreds of diatom genes were differentially expressed in the iron-enriched community when compared to the iron-limited community; transcripts of diatom genes required for synthesis of photosynthesis and chlorophyll components, nitrate assimilation and the urea cycle, and synthesis of carbohydrate storage compounds were significantly over-represented. Transcripts of genes encoding rhodopsins in eukaryotic phytoplankton were significantly under-represented following iron enrichment, suggesting rhodopsins help cells cope with low-iron conditions. Oceanic diatoms appear to display a distinctive transcriptional response to iron enrichment that allows chemical reduction of available nitrogen and carbon sources along with a continued dependence on iron-free photosynthetic proteins rather than substituting for iron-containing functional equivalents present within their gene repertoire. This ability of diatoms to divert their newly acquired iron towards nitrate assimilation may underlie why diatoms consistently dominate iron enrichments in HNLC regions.
Additional links discussing our findings:
1) Recommended Paper – Faculty of 1000
Data archive can be found here.
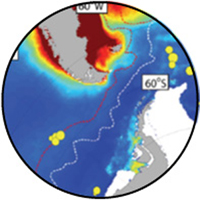
 Iron and silicic acid effects on phytoplankton in the central equatorial Pacific Ocean
Iron and silicic acid effects on phytoplankton in the central equatorial Pacific Ocean
The Central Equatorial Pacific (CEP) has received much attention by oceanographers as it is a well-characterized iron-limited region and has the largest global efflux of CO2 from the ocean to the atmosphere due to the continuous upwelling of CO2-rich water. Currently, there is debate as to whether low iron and low Si(OH)4 concentrations co-regulate diatom growth and/or silicic acid uptake in this region. We performed microcosm nutrient amendment experiments in the CEP investigating the influence of Fe and Si(OH)4 additions on phytoplankton by assessing Si, C and N utilization as well as the resulting changes in the species diversity and chemical compositions following these nutrient amendments. Our results show that the addition of Fe alone enhances diatom productivity but that Si(OH)4 concentrations limits the extent to which large diatoms can form their silica-containing frustules, restricting cell division but not their ability to assimilate C and N. In contrast, the addition of Fe and Si(OH)4 results in a higher abundance of large diatoms that are also more heavily silicified. The measured variability in the iron-replete diatoms’ chemical and biochemical compositions as a function of Si nutritional status may have a considerable influence on their effectiveness as exporters of C from mixed-layer waters to the ocean’s depths.
 The morphometrics of pennate diatom frustules as potential indicators of iron-limited growth in past oceans
The morphometrics of pennate diatom frustules as potential indicators of iron-limited growth in past oceans
Diatom growth and nutrient utilization in large regions of the world’s oceans are regulated by iron availability. Under iron limitation, both centric and pennate forms of diatoms decrease in size. The associated increase in surface area-to-volume (SA:V) ratio may improve nutrient uptake kinetics. In parallel, cellular Si concentrations are elevated in iron-limited diatoms relative to N and C. Variations in nutritional requirements of diatoms acclimated to low-iron conditions have been hypothesized to account for higher cellular Si or lower cellular N and C. Changes in the Si-containing valve surface area relative to volume in some diatoms may also account for variations in the cellular Si:N and Si:C ratios. In particular, iron-limited pennate diatoms of the genus Pseudo-nitzschia have reduced widths relative to their lengths (i.e. lower length normalized widths, LNW) compared to iron-replete cells. In the pennate diatom Fragilariopsis kerguelensis, the mean LNWs of valves preserved in sediments throughout the Southern Ocean (a well-characterized iron-limited region) is positively correlated to satellite-derived, climatological net primary productivity in the overlying waters. Because of the specific morphological changes in pennate diatom frustules in response to iron availability, we propose that the physical properties (e.g. LNWs) of frustules preserved in the sediments can be a valuable paleoceanographic proxy for Fe-limited diatom growth.
Iron storage by ferritin in  diatoms
diatoms
We recently discovered that the bloom-forming pennate diatoms Pseudo-nitzschia and Fragilariopsis use the iron-concentrating protein, ferritin, to safely store iron. This is the first description of ferritin in any member of the Stramenopiles, a diverse Eukaryotic lineage that includes unicellular algae, macroalgae and plant parasites. Phylogenetic analyses suggest that ferritin may have arisen in this small subset of diatoms via a lateral gene transfer. The crystal structure and functional assays of recombinant ferritin derived from Pseudo-nitzschia multiseries reveal a maxi-ferritin that exhibits ferroxidase activity and binds iron. The protein is predicted to be targeted to the chloroplast to control the distribution and storage of iron for proper functioning of the photosynthetic machinery. Abundance of Pseudo-nitzschia ferritin transcripts is regulated by iron nutritional status and is closely tied to the loss and recovery of photosynthetic competence. Enhanced iron storage with ferritin allows the oceanic diatom Pseudo-nitzschia granii to undergo several more cell divisions in the absence of iron than the comparably sized, oceanic centric diatom Thalassiosira oceanica. Ferritin in pennate diatoms likely contributes to their success in chronically low iron regions that receive intermittent iron inputs and provides an explanation for the importance of these organisms in regulating oceanic CO2 over geological timescales.
Additional links discussing our findings:
1) Recommended Paper – Faculty of 1000
2) Putting the Bloom on the Diatom – Canadian Light Source Inc.
3) Marine Diatoms Survive Iron Droughts in the Ocean by Storing Iron in Ferritin – Stanford Synchrotron Radiation Lightsource

University Operator: (919) 962-2211 | © 2024 The University of North Carolina at Chapel Hill |